This is a wrapper for the psych functions psych::pca() and psych::fa()
to produce output that it similar to the output produced by jamovi.
factorAnalysis(
data,
nfactors,
items = names(data),
rotate = "oblimin",
covar = FALSE,
na.rm = TRUE,
kaiser = 1,
loadings = TRUE,
summary = FALSE,
correlations = FALSE,
modelFit = FALSE,
eigenValues = FALSE,
screePlot = FALSE,
residuals = FALSE,
itemLabels = items,
colorLoadings = FALSE,
fm = "minres",
digits = 2,
headingLevel = 3,
...
)
principalComponentAnalysis(
data,
items,
nfactors,
rotate = "oblimin",
covar = FALSE,
na.rm = TRUE,
kaiser = 1,
loadings = TRUE,
summary = FALSE,
correlations = FALSE,
eigenValues = FALSE,
screePlot = FALSE,
residuals = FALSE,
itemLabels = items,
colorLoadings = FALSE,
digits = 2,
headingLevel = 3,
...
)
rosettaDataReduction_partial(
x,
digits = x$input$digits,
headingLevel = x$input$headingLevel,
echoPartial = FALSE,
partialFile = NULL,
quiet = TRUE,
...
)
# S3 method for rosettaDataReduction
knit_print(
x,
digits = x$input$digits,
headingLevel = x$input$headingLevel,
echoPartial = FALSE,
partialFile = NULL,
quiet = TRUE,
...
)
# S3 method for rosettaDataReduction
print(
x,
digits = x$input$digits,
headingLevel = x$input$headingLevel,
forceKnitrOutput = FALSE,
...
)Arguments
- data
The data frame that contains the
items.- nfactors
The number of factors to extract, or '
eigen' to extract all factors with an eigen value higher than the number specified inkaiser. In the future,parallelcan be specified here to extract the number of factors suggested by parallel analysis.- items
The items to analyse; if not specified, all variables in
datawill be used.- rotate
Which rotation to use; see
psych::fa()for all options. The most common options are 'none' to not rotate at all; 'varimax' for an orthogonal rotation (assuming/imposing that the components or factors are not correlated); or 'oblimin' for an oblique rotation (allowing the components/factors to correlate).- covar
Whether to analyse the correlation matrix (
FALSE) or the covariance matrix (TRUE).- na.rm
Whether to first remove all cases with missing values.
- kaiser
The minimum eigenvalue when applying the Kaiser criterion (see
nfactors).- loadings
Whether to display the component or factor loadings.
- summary
Whether to display the factor or component summary.
- correlations
Whether to display the correlations between factors of components.
- modelFit
Whether to display the model fit Only for EFA).
- eigenValues
Whether to display the eigen values.
- screePlot
Whether to display the scree plot.
- residuals
Whether to display the matrix with residuals.
- itemLabels
Optionally, labels to use for the items (optionally, named, with the names corresponding to the
items; otherwise, the order of the labels has to match the order of the items)- colorLoadings
Whether, when producing an Rmd partial (i.e. when calling the command while knitting) to colour the cells using
kableExtra::kable_styling().- fm
The method to use for the factor analysis: '
fm' for Minimum Residuals; 'ml' for Maximum Likelihood; and 'pa' for Principal Factor.- digits
The number of digits to round to.
- headingLevel
The number of hashes to print in front of the headings when printing while knitting
- ...
Any additional arguments are passed to
psych::fa(),psych::pca(), to the default print method by the print method, and tormdpartials::partial()when knitting an RMarkdown partial.- x
The object to print.
- echoPartial
Whether to show the executed code in the R Markdown partial (
TRUE) or not (FALSE).- partialFile
This can be used to specify a custom partial file. The file will have object
xavailable.- quiet
Passed on to
knitr::knit()whether it should b chatty (FALSE) or quiet (TRUE).- forceKnitrOutput
Force knitr output.
Value
An object with the object resulting from the call to the
psych functions and some extracted information that will be printed.
Details
The code in these functions uses parts of the code in jamovi, written by Jonathon Love and Ravi Selker.
Examples
### Load example dataset
data("pp15", package="rosetta");
### Get variable names with expected
### effects of a high dose of MDMA
items <-
grep(
"highDose_AttBeliefs_",
names(pp15),
value=TRUE
);
### Do a factor analysis
rosetta::factorAnalysis(
data = pp15,
items = items,
nfactors = "eigen",
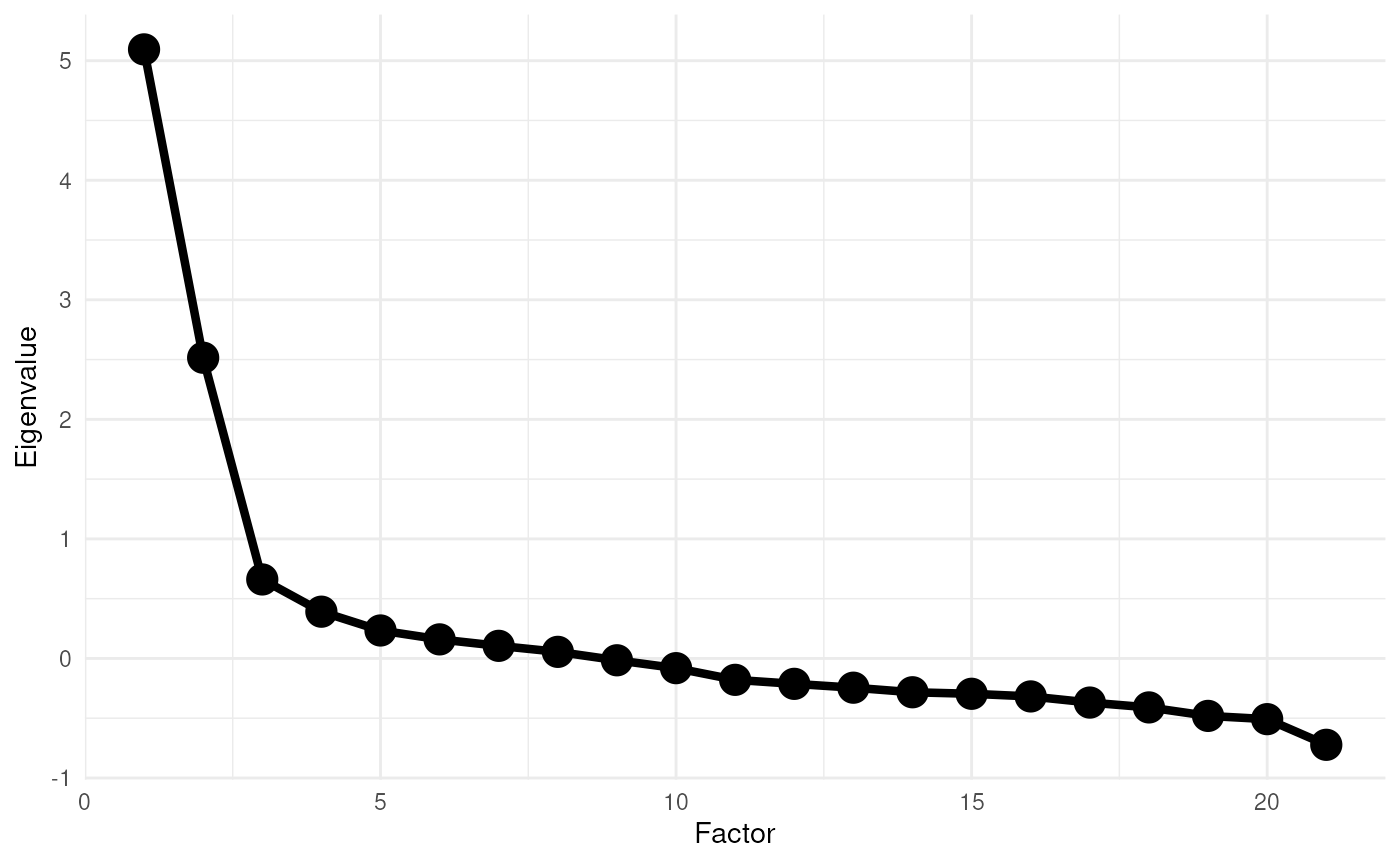
scree = TRUE
);
#>
#> Exploratory Factor Analysis (EFA)
#>
#> Extraction method: Minimum Residuals
#> Rotation : Oblimin rotation
#> Sample size : 212
#>
#> Factor loadings
#>
#> Factor 1 Factor 2 Uniqueness
#> highDose_AttBeliefs_long 0.24 0.37 0.82
#> highDose_AttBeliefs_intensity 0.16 0.73 0.45
#> highDose_AttBeliefs_intoxicated 0.02 0.69 0.52
#> highDose_AttBeliefs_energy 0.45 0.11 0.79
#> highDose_AttBeliefs_euphoria 0.73 0.23 0.44
#> highDose_AttBeliefs_insight 0.69 -0.14 0.50
#> highDose_AttBeliefs_connection 0.77 0.12 0.40
#> highDose_AttBeliefs_contact 0.77 0.09 0.40
#> highDose_AttBeliefs_sex 0.41 -0.16 0.80
#> highDose_AttBeliefs_coping 0.18 0.47 0.75
#> highDose_AttBeliefs_isolated -0.52 0.14 0.71
#> highDose_AttBeliefs_boundaries 0.31 -0.09 0.89
#> highDose_AttBeliefs_music 0.56 0.36 0.58
#> highDose_AttBeliefs_hallucinate -0.24 0.47 0.72
#> highDose_AttBeliefs_timeAwareness 0.01 0.54 0.71
#> highDose_AttBeliefs_memory 0.40 -0.40 0.66
#> highDose_AttBeliefs_health 0.34 -0.51 0.60
#> highDose_AttBeliefs_better 0.71 -0.05 0.49
#> highDose_AttBeliefs_physicalSideEffects -0.46 0.38 0.63
#> highDose_AttBeliefs_psychicSideEffects -0.53 0.30 0.61
#> highDose_AttBeliefs_regret -0.55 0.31 0.58
#> <table style='border:0px solid black !important; font-family: "Arial Narrow", "Source Sans Pro", sans-serif; margin-left: auto; margin-right: auto;' class="table table-condensed">
#> <thead>
#> <tr>
#> <th style="text-align:left;"> </th>
#> <th style="text-align:right;"> Factor 1 </th>
#> <th style="text-align:right;"> Factor 2 </th>
#> <th style="text-align:right;"> Uniqueness </th>
#> </tr>
#> </thead>
#> <tbody>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_long </td>
#> <td style="text-align:right;"> 0.24 </td>
#> <td style="text-align:right;"> 0.37 </td>
#> <td style="text-align:right;"> 0.82 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_intensity </td>
#> <td style="text-align:right;"> 0.16 </td>
#> <td style="text-align:right;"> 0.73 </td>
#> <td style="text-align:right;"> 0.45 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_intoxicated </td>
#> <td style="text-align:right;"> 0.02 </td>
#> <td style="text-align:right;"> 0.69 </td>
#> <td style="text-align:right;"> 0.52 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_energy </td>
#> <td style="text-align:right;"> 0.45 </td>
#> <td style="text-align:right;"> 0.11 </td>
#> <td style="text-align:right;"> 0.79 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_euphoria </td>
#> <td style="text-align:right;"> 0.73 </td>
#> <td style="text-align:right;"> 0.23 </td>
#> <td style="text-align:right;"> 0.44 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_insight </td>
#> <td style="text-align:right;"> 0.69 </td>
#> <td style="text-align:right;"> -0.14 </td>
#> <td style="text-align:right;"> 0.50 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_connection </td>
#> <td style="text-align:right;"> 0.77 </td>
#> <td style="text-align:right;"> 0.12 </td>
#> <td style="text-align:right;"> 0.40 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_contact </td>
#> <td style="text-align:right;"> 0.77 </td>
#> <td style="text-align:right;"> 0.09 </td>
#> <td style="text-align:right;"> 0.40 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_sex </td>
#> <td style="text-align:right;"> 0.41 </td>
#> <td style="text-align:right;"> -0.16 </td>
#> <td style="text-align:right;"> 0.80 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_coping </td>
#> <td style="text-align:right;"> 0.18 </td>
#> <td style="text-align:right;"> 0.47 </td>
#> <td style="text-align:right;"> 0.75 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_isolated </td>
#> <td style="text-align:right;"> -0.52 </td>
#> <td style="text-align:right;"> 0.14 </td>
#> <td style="text-align:right;"> 0.71 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_boundaries </td>
#> <td style="text-align:right;"> 0.31 </td>
#> <td style="text-align:right;"> -0.09 </td>
#> <td style="text-align:right;"> 0.89 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_music </td>
#> <td style="text-align:right;"> 0.56 </td>
#> <td style="text-align:right;"> 0.36 </td>
#> <td style="text-align:right;"> 0.58 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_hallucinate </td>
#> <td style="text-align:right;"> -0.24 </td>
#> <td style="text-align:right;"> 0.47 </td>
#> <td style="text-align:right;"> 0.72 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_timeAwareness </td>
#> <td style="text-align:right;"> 0.01 </td>
#> <td style="text-align:right;"> 0.54 </td>
#> <td style="text-align:right;"> 0.71 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_memory </td>
#> <td style="text-align:right;"> 0.40 </td>
#> <td style="text-align:right;"> -0.40 </td>
#> <td style="text-align:right;"> 0.66 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_health </td>
#> <td style="text-align:right;"> 0.34 </td>
#> <td style="text-align:right;"> -0.51 </td>
#> <td style="text-align:right;"> 0.60 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_better </td>
#> <td style="text-align:right;"> 0.71 </td>
#> <td style="text-align:right;"> -0.05 </td>
#> <td style="text-align:right;"> 0.49 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_physicalSideEffects </td>
#> <td style="text-align:right;"> -0.46 </td>
#> <td style="text-align:right;"> 0.38 </td>
#> <td style="text-align:right;"> 0.63 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_psychicSideEffects </td>
#> <td style="text-align:right;"> -0.53 </td>
#> <td style="text-align:right;"> 0.30 </td>
#> <td style="text-align:right;"> 0.61 </td>
#> </tr>
#> <tr>
#> <td style="text-align:left;"> highDose_AttBeliefs_regret </td>
#> <td style="text-align:right;"> -0.55 </td>
#> <td style="text-align:right;"> 0.31 </td>
#> <td style="text-align:right;"> 0.58 </td>
#> </tr>
#> </tbody>
#> </table>
 if (FALSE) {
### To get more output, show the
### output as Rmd Partial in the viewer,
### and color/size the factor loadings
rosetta::rosettaDataReduction_partial(
rosetta::factorAnalysis(
data = pp15,
items = items,
nfactors = "eigen",
summary = TRUE,
correlations = TRUE,
colorLoadings = TRUE
)
);
}
if (FALSE) {
### To get more output, show the
### output as Rmd Partial in the viewer,
### and color/size the factor loadings
rosetta::rosettaDataReduction_partial(
rosetta::factorAnalysis(
data = pp15,
items = items,
nfactors = "eigen",
summary = TRUE,
correlations = TRUE,
colorLoadings = TRUE
)
);
}